SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study
Abstract
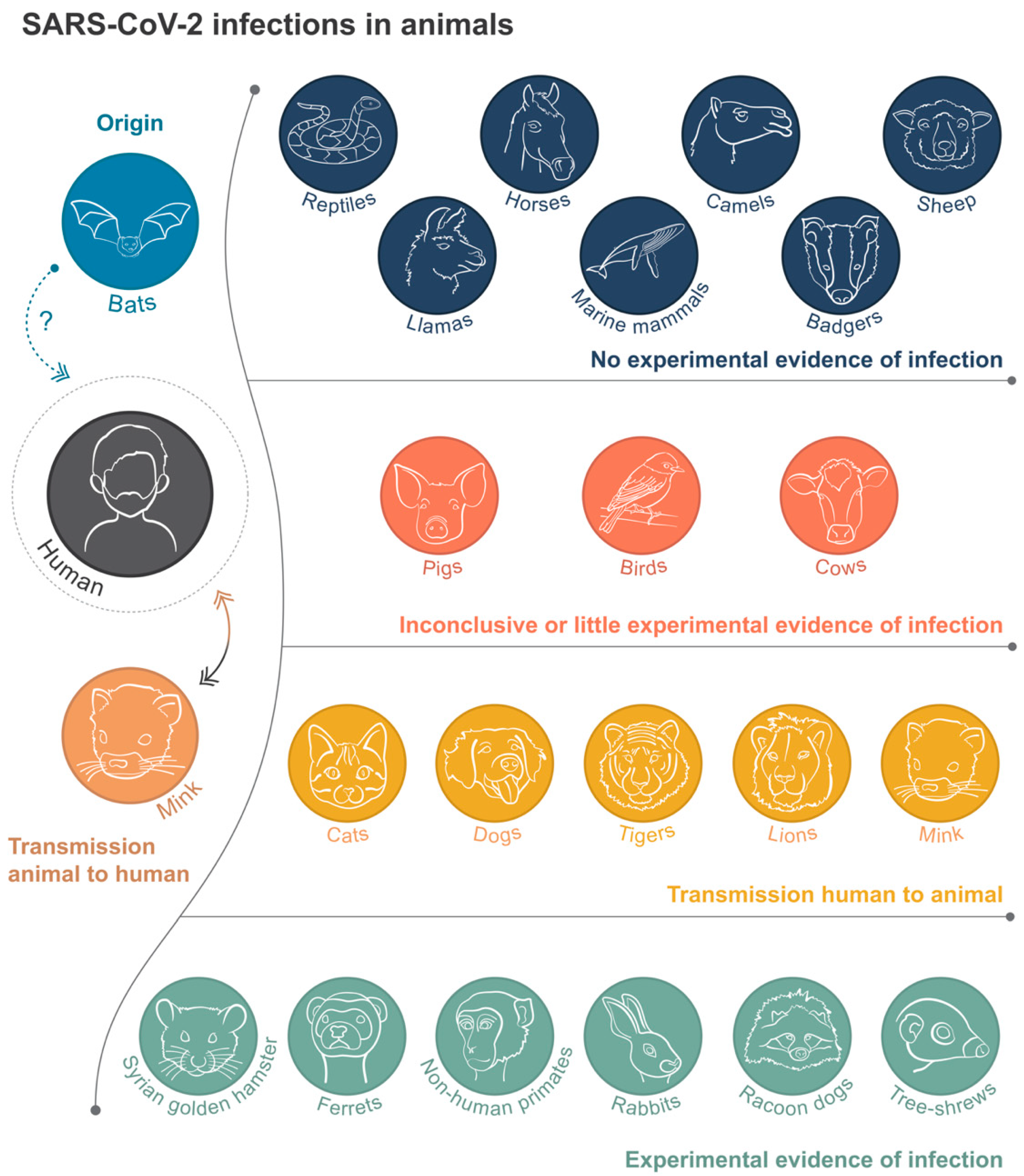
1. Introduction
2. SARS-CoV-2 Infections in Animals
2.1. In the Wild
2.2. Livestock Farming
2.3. Zoos
2.4. Companion Animals
2.4.1. Cats
2.4.2. Dogs and Other Pets
3. Further Experimental Proof of Permissiveness to Infection
4. Experimental Models of COVID-19 Disease
5. Areas for Further Investigation
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Wang, L.F.; Eaton, B.T. Bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007, 315, 325–344. [Google Scholar] [CrossRef]
- Dudas, G.; Carvalho, L.M.; Rambaut, A.; Bedford, T. MERS-CoV spillover at the camel-human interface. Elife 2018, 7. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, 127–147. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e3. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2. [Google Scholar] [CrossRef]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, J.; Zhu, J.; Ding, X.; Lan, T.; Zhu, L.; Xiang, R.; Ding, P.; Wang, H.; Wang, X.; et al. Single-cell screening of SARS-CoV-2 target cells in pets, livestock, poultry and wildlife. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lee, J.; Hughes, T.; Lee, M.H.; Field, H.; Rovie-Ryan, J.J.; Sitam, F.T.; Sipangkui, S.; Nathan, S.K.; Ramirez, D.; Kumar, S.V.; et al. No evidence of coronaviruses or other potentially zoonotic viruses in Sunda pangolins (Manis javanica) entering the wildlife trade via Malaysia. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Ulrich, L. Experimental infection of cattle with SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Gultom, M.; Licheri, M.; Laloli, L.; Wider, M.; Straessle, M.; Steiner, S.; Kratzel, A.; Thao, T.T.; Stalder, H.; Portmann, J.; et al. Susceptibility of well-differentiated airway epithelial cell 1 cultures from domestic and wildlife animals to SARS-CoV-2 2 3. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cahan, E. COVID-19 hits U.S. mink farms after ripping through Europe. Science 2020. [Google Scholar] [CrossRef]
- OIE—World Organisation for Animal Health. Events in Animals. 2020. Available online: https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/events-in-animals/ (accessed on 28 August 2020).
- Rabalski, L.; Kosinski, M.; Smura, T.; Aaltonen, K.; Kant, R.; Szewczyk, B.; Grzybek, M. Detection and molecular characterisation of SARS-CoV-2 1 in farmed mink (Neovision vision) in Poland 2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Munnink, B.B.; Hakze-van Der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef] [PubMed]
- Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; Van Der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2020, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M. Coronavirus rips through Dutc mink farms, triggering culls. Science 2020, 368, 1169. [Google Scholar] [CrossRef]
- Van Dorp, L.; Tan, C.C.; Lam, S.D.; Richard, D.; Owen, C.; Berchtold, D.; Orengo, C.; Balloux, F. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv 2020. [Google Scholar] [CrossRef]
- WHO. SARS-CoV-2 Mink-Associated Variant Strain—Denmark. 2020. Available online: https://www.who.int/csr/don/06-november-2020-mink-associated-sars-cov2-denmark/en/ (accessed on 10 November 2020).
- ECDC. Detection of New SARS-CoV-2 Variants Related to Mink. Eur. Cent. Dis. Prev. Control. 12 November 2020. ECDC: Stockholm. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf (accessed on 7 January 2021).
- WHO. Coronavirus Disease (COVID-19) Outbreak—Mink-Strain of COVID-19 Virus in Denmark. 2020. Available online: https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/11/mink-strain-of-covid-19-virus-in-denmark (accessed on 10 November 2020).
- GOV.UK. New SARS-CoV-2 Variant—GOV.UK. 2020. Available online: https://www.gov.uk/government/collections/new-sars-cov-2-variant (accessed on 7 January 2021).
- Mallapaty, S. COVID mink analysis shows mutations are not dangerous—Yet. Nature 2020, 587, 340–341. [Google Scholar] [CrossRef]
- Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Jumping back and forth: Anthropozoonotic and zoonotic transmission of SARS-CoV-2 on mink farms Affiliations. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.; et al. Susceptibility of rabbits to SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, L.; Mitchell, P.K.; Calle, P.P.; Bartlett, S.L.; McAloose, D.; Killian, M.L.; Yuan, F.; Fang, Y.; Goodman, L.B.; Fredrickson, R.; et al. 2020 Complete Genome Sequence of SARS-CoV-2 in a Tiger from a U.S. Zoological Collection. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- USDA. USDA Statement on the Confirmation of COVID-19 in a Tiger in New York. 2020. Available online: https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/ny-zoo-covid-19 (accessed on 12 August 2020).
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From people to Panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bartlett, S.L.; Diel, D.G.; Wang, L.; Zec, S.; Laverack, M.; Martins, M.; Caserta, L.C.; Killian, M.L.; Terio, K.; Olmstead, C.; et al. SARS-CoV-2 Infection and Longitudinal Fecal Screening In Malayan Tigers (Panthera tigris jacksoni), Amur Tigers (Panthera tigris altaica), And African Lions (Panthera leo krugeri) At The Bronx Zoo, New York, USA. bioRxiv 2020. [Google Scholar] [CrossRef]
- USDA APHIS. Confirmation of COVID-19 in Gorillas at a California Zoo. 2021. Available online: https://content.govdelivery.com/accounts/USDAAPHIS/bulletins/2b5837f/ (accessed on 13 January 2021).
- Wang, M.; Jing, H.Q.; Xu, H.F.; Jiang, X.G.; Kan, B.; Liu, Q.Y.; Wan, K.L.; Cui, B.Y.; Zheng, H.; Cui, Z.G.; et al. Surveillance on severe acute respiratory syndrome associated coronavirus in animals at a live animal market of Guangzhou in 2004. Zhonghua Liu Xing Bing Xue Za Zhi 2005, 26, 84–87. [Google Scholar] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; Zhao, Y. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats Running title: SARS-CoV-2 in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.A.; Pauvolid-Corrêa, A.; Zecca, I.B.; Davila, E.; Auckland, L.D.; Roundy, C.M.; Tang, W.; Torchetti, M.; Killian, M.L.; Jenkins-Moore, M.; et al. Natural SARS-CoV-2 infections, including virus isolation, among serially tested cats and dogs in households with confirmed human COVID-19 cases in Texas, USA. bioRxiv 2020. [Google Scholar] [CrossRef]
- USDA APHIS. Confirmed Cases of SARS-CoV-2 in Animals in the United States. 2020. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_one_health/sars-cov-2-animals-us (accessed on 28 August 2020).
- Musso, N.; Costantino, A.; La Spina, S.; Finocchiaro, A.; Andronico, F.; Stracquadanio, S.; Liotta, L.; Visalli, R.; Emmanuele, G. New SARS-CoV-2 Infection Detected in an Italian Pet Cat by RT-qPCR from Deep Pharyngeal Swab. Pathogens 2020, 9, 746. [Google Scholar] [CrossRef]
- Barrs, V.R.; Peiris, M.; Tam, K.W.; Law, P.Y.; Brackman, C.J.; To, E.M.; Yu, V.Y.; Chu, D.K.; Perera, R.A.; Sit, T.H. SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China. Emerg. Infect. Dis. 2020, 26, 3071–3074. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Dumarest, M.; Vanhomwegen, J.; Delaplace, M.; Caro, V.; Kwasiborski, A.; Hourdel, V.; Chevaillier, P.; Barbarino, A.; Comtet, L.; et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibáñez, S.; Santibáñez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Puig, M.; Rodon, J.; Avila-Nieto, C.; Carrillo, J.; Cantero, G.; Terrón, M.T.; Cruz, S.; Parera, M.; Noguera-Julián, M.; et al. Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. USA 2020, 117, 24790–24793. [Google Scholar] [CrossRef]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Respiratory disease in cats associated with human-to-cat transmission of SARS-CoV-2 in the UK. bioRxiv 2020. [Google Scholar] [CrossRef]
- Neira, V.; Brito, B.; Agüero, B.; Berrios, F.; Valdés, V.; Gutierrez, A.; Ariyama, N.; Espinoza, P.; Retamal, P.; Holmes, E.C.; et al. A household case evidences shorter shedding of SARS-CoV-2 in naturally infected cats 6 compared to their human owners. 7 Affiliations. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Jin, Y.; Liu, Y.; Sun, J.; Hao, L.; Bai, J.; Huang, T.; Lin, D.; Jin, Y.; Tian, K. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 2020, 67, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liu, Y.; Sun, C.; Bai, J.; Sun, J.; Hao, L.; Li, X.; Tian, K. SARS-CoV-2 Serological Survey of Cats in China before and after the Pandemic. Virol. Sin. 2020, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Barbarino, A.; Maso, D.; Behillil, S.; Enouf, V.; Huon, C.; Jaraud, A.; Chevallier, L.; Backovic, M.; Pérot, P.; et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health 2020, 10, 100164. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2020, 11, 100192. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Carossino, M.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; Madden, D.W.; Cool, K.; Artiaga, B.L.; McDowell, C.; Bold, D.; et al. Experimentally re-infected cats do not transmit SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sit, T.H.; Brackman, C.J.; Ip, S.M.; Tam, K.W.; Law, P.Y.; To, E.M.; Veronica, Y.T.; Sims, L.D.; Tsang, D.N.; Chu, D.K.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Delong, J. Dutch Minister Confirms Dog, Three Cats Have Caught Novel Coronavirus|Armenian American Reporter. 2020. Available online: https://www.reporter.am/dutch-minister-confirms-dog-three-cats-have-caught-novel-coronavirus/ (accessed on 17 August 2020).
- Perisé-Barrios, A.J.; Tomeo-Martín, B.D.; Gómez-Ochoa, P.; Delgado-Bonet, P.; Plaza, P.; Palau-Concejo, P.; González, J.; Ortiz-Díez, G.; Meléndez-Lazo, A.; Gentil, M.; et al. Humoral response to SARS-CoV-2 by healthy and sick dogs during COVID-19 pandemic in Spain. bioRxiv 2020. [Google Scholar] [CrossRef]
- McNamara, T.; Richt, J.A.; Glickman, L. A Critical Needs Assessment for Research in Companion Animals and Livestock Following the Pandemic of COVID-19 in Humans. Vector-Borne Zoonotic Dis. 2020, 20, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Fagre, A.; Lewis, J.; Eckley, M.; Zhan, S.; Rocha, S.M.; Sexton, N.R.; Burke, B.; Geiss, B.; Peersen, O.; Kading, R.; et al. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for reverse zoonosis to New World rodents. bioRxiv 2020. [Google Scholar] [CrossRef]
- Griffin, B.D.; Chan, M.; Tailor, N.; Mendoza, E.J.; Leung, A.; Warner, B.M.; Duggan, A.T.; Moffat, E.; He, S.; Garnett, L.; et al. North American deer mice are susceptible to SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Pickering, B.S.; Smith, G.; Pinette, M.M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Suarez, D.L.; Pantin-Jackwood, M.J.; Swayne, D.E.; Lee, S.A.; Deblois, S.M.; Spackman, E. Lack of susceptibility of poultry to SARS-CoV-2 and MERS-CoV. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Freuling, C.M.; Breithaupt, A.; Müller, T.; Sehl, J.; Balkema-Buschmann, A.; Rissmann, M.; Klein, A.; Wylezich, C.; Höper, D.; Wernike, K.; et al. 2020 Susceptibility of raccoon dogs for SARS-CoV-2 Title: Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, J.; Kuang, D.; Xu, J.; Yang, M.; Ma, C.; Zhao, S.; Li, J.; Long, H.; Ding, K.; et al. 2020 Susceptibility of tree shrew to SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.C.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ge, X.Y.; Yang, W.H.; Zhou, J.H.; Li, B.; Zhang, W.; Shi, Z.L.; Zhang, Y.Z. Detection of alpha- and betacoronaviruses in rodents from Yunnan, China. Virol. J. 2017, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Clark, J.J.; Penrice-Randal, R.; Sharma, P.; Kipar, A.; Dong, X.; Davidson, A.D.; Williamson, M.K.; Matthews, D.A.; Turtle, L.; Prince, T.; et al. Sequential infection with influenza A virus followed by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) leads to more severe disease and encephalitis in a mouse model of COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jiang, R.D.; Liu, M.Q.; Chen, Y.; Shan, C.; Zhou, Y.W.; Shen, X.R.; Li, Q.; Zhang, L.; Zhu, Y.; Si, H.R.; et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 2020, 182, 50–58.e8. [Google Scholar] [CrossRef] [PubMed]
- Leist, S.R.; Dinnon, I.I.I.K.H.; Schäfer, A.; Longping, V.T.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-adapted SARS-CoV-2 induces Acute Lung Injury (ALI) and mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020, 71, 2428–2446. [Google Scholar] [CrossRef] [PubMed]
- Bertzbach, L.D.; Vladimirova, D.; Dietert, K.; Abdelgawad, A.; Gruber, A.D.; Osterrieder, N.; Trimpert, J. SARS-CoV-2 infection of Chinese hamsters (Cricetulus griseus) reproduces COVID-19 pneumonia in a well-established small animal model. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Kok, A.; de Meulder, D.; Bestebroer, T.M.; Lamers, M.M.; Okba, N.M.; van Vlissingen, M.F.; Rockx, B.; Haagmans, B.L.; Koopmans, M.P.; et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020, 11, 1–6. [Google Scholar] [CrossRef]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709.e2. [Google Scholar] [CrossRef] [PubMed]
- Kutter, J.S.; de Meulder, D.; Bestebroer, T.M.; Lexmond, P.; Fouchier, R.A.; Herfst, S. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sawatzki, K.; Hill, N.; Puryear, W.; Foss, A.; Stone, J. Ferrets not infected by SARS-CoV-2 in a high-exposure domestic setting. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Gao, H.; Xiao, C.; Liu, J.; Xue, J.; Lv, Q.; Liu, J.; Yu, P.; Xu, Y.; et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chandrashekar, A.; Liu, J.; Martinot, A.J.; McMahan, K.; Mercado, N.B.; Peter, L.; Tostanoski, L.H.; Yu, J.; Maliga, Z.; Nekorchuk, M.; et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020, 369, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef] [PubMed]
- Woolsey, C.; Borisevich, V.; Prasad, A.N.; Agans, K.N.; Deer, D.J.; Dobias, N.S.; Heymann, J.C.; Foster, S.L.; Levine, C.B.; Medina, L.; et al. Establishment of an African green monkey model for COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cross, R.W.; Agans, K.N.; Prasad, A.N.; Borisevich, V.; Woolsey, C.; Deer, D.J.; Dobias, N.S.; Geisbert, J.B.; Fenton, K.A.; Geisbert, T.W. Intranasal exposure of African green monkeys to SARS-CoV-2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol. J. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; Ma, C.; et al. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Stoddart, A.K.; Gagnon, G.A.; Dellaire, G. Pandemic danger to the deep: The risk of marine mammals contracting SARS-CoV-2 from wastewater. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Hewson, I. Coronaviruses in the Sea. Front. Microbiol. 2020, 11, 1795. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Atoni, E.; Zhao, L.; Ren, N.; Huang, D.; Pei, R.; Chen, Z.; Xiong, J.; Nyaruaba, R.; Xiao, S.; et al. SARS-CoV-2 Does Not Replicate in Aedes Mosquito Cells nor Present in Field-Caught Mosquitoes from Wuhan. Virol. Sin. 2020, 35, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.S.; Vanlandingham, D.L.; Bilyeu, A.N.; Sharp, H.M.; Hettenbach, S.M.; Higgs, S. SARS-CoV-2 failure to infect or replicate in mosquitoes: An extreme challenge. Sci. Rep. 2020, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Balaraman, V.; Drolet, B.S.; Gaudreault, N.N.; Wilson, W.C.; Owens, J.; Bold, D.; Swanson, D.A.; Jasperson, D.C.; Noronha, L.E.; Richt, J.A.; et al. Susceptibility of midge and mosquito vectors to SARS-CoV-2 by natural route of infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dai, M.; Li, H.; Yan, N.; Huang, J.; Zhao, L.; Xu, S.; Jiang, S.; Pan, C.; Liao, M. Long-term survival of salmon-attached SARS-CoV-2 at 4 °C as a potential source of 2 transmission in seafood markets Running Title: Survival of salmon-attached SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- 2021 COVID-19: Ice Cream Tests Positive for Coronavirus in China|World News|Sky News. Available online: https://news.sky.com/story/covid-19-ice-cream-tests-positive-for-coronavirus-in-china-12188761 (accessed on 18 January 2021).
- Fisher, D.; Reilly, A.; Kang, A.; Zheng, E.; Cook, A.R.; Anderson, D.E. Seeding of outbreaks of COVID-19 by contaminated fresh and frozen food. bioRxiv 2020. [Google Scholar] [CrossRef]
- Waltenburg, M.A.; Victoroff, T.; Rose, C.E.; Butterfield, M.; Jervis, R.H.; Fedak, K.M.; Gabel, J.A.; Feldpausch, A.; Dunne, E.M.; Austin, C.; et al. Update: COVID-19 Among Workers in Meat and Poultry Processing Facilities—United States, April–May 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Sarvikivi, E.; Roivainen, M.; Maunula, L.; Niskanen, T.; Korhonen, T.; Lappalainen, M.; Kuusi, M. Multiple norovirus outbreaks linked to imported frozen raspberries. Epidemiol. Infect. 2012, 140, 260–267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kegg, D.; Gluckman, P.; Boulton, G.; Hackmann, H.; Karim, S.S.A.; Piot, P.; Woopen, C. Future scenarios for the COVID-19 pandemic. Lancet 2021, 397, 777–778. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prince, T.; Smith, S.L.; Radford, A.D.; Solomon, T.; Hughes, G.L.; Patterson, E.I. SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study. Viruses 2021, 13, 494. https://doi.org/10.3390/v13030494
Prince T, Smith SL, Radford AD, Solomon T, Hughes GL, Patterson EI. SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study. Viruses. 2021; 13(3):494. https://doi.org/10.3390/v13030494
Chicago/Turabian StylePrince, Tessa, Shirley L. Smith, Alan D. Radford, Tom Solomon, Grant L. Hughes, and Edward I. Patterson. 2021. "SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study" Viruses 13, no. 3: 494. https://doi.org/10.3390/v13030494
APA StylePrince, T., Smith, S. L., Radford, A. D., Solomon, T., Hughes, G. L., & Patterson, E. I. (2021). SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study. Viruses, 13(3), 494. https://doi.org/10.3390/v13030494





